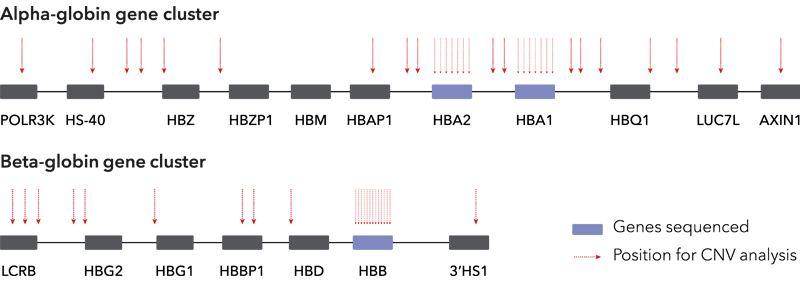
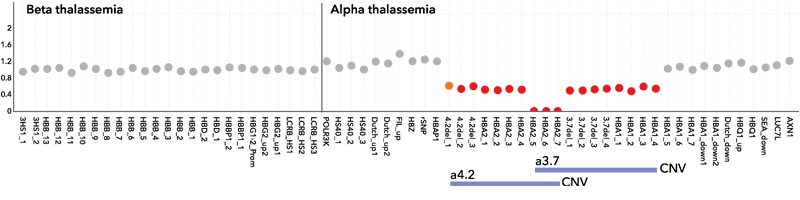
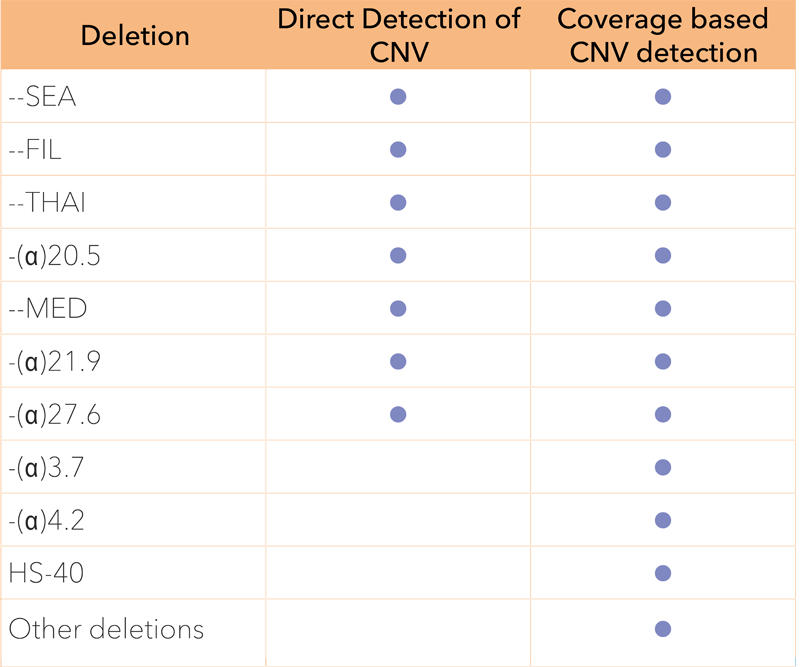
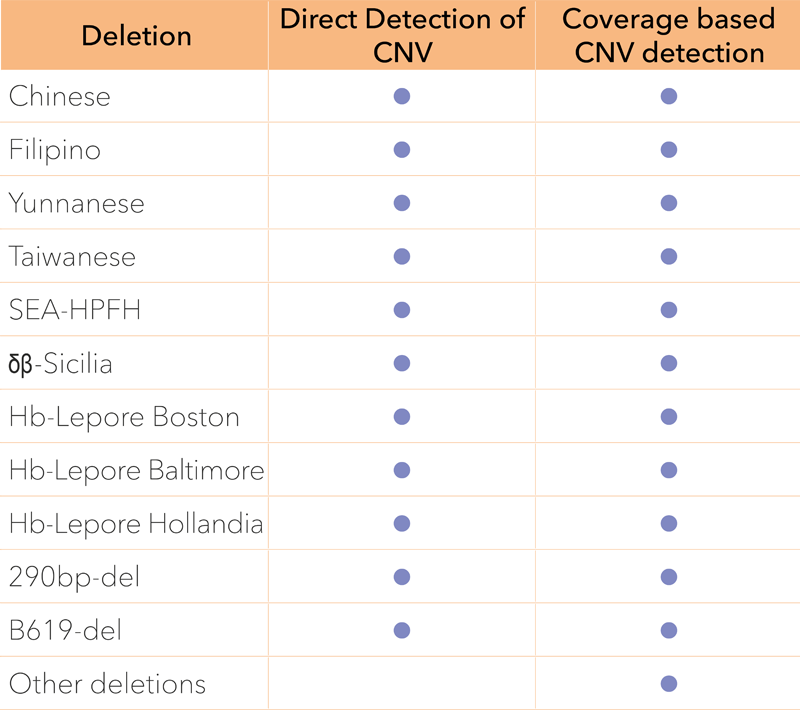
A single, one-tube NGS assay that detects all sequence variants in HBA1, HBA2 and HBB in a single run, eliminating the need for additional workflows. The assay detects SNVs, Indels and applies two simultaneous methods to detect CNVs. The extremely simple procedure suits any lab, whether you are running advanced genetic testing or large scale mutation screening.
-
 Complete single-tube analysis for thalassemias
Complete single-tube analysis for thalassemias Fast and simple NGS workflow
Fast and simple NGS workflow User-friendly data analysis software
User-friendly data analysis software