VarSome Clinical is a clinically-certified platform allowing fast and accurate variant discovery, annotation, and interpretation of NGS data for whole genomes, exomes, and gene panels. VarSome Clinical helps molecular geneticists and clinicians reach faster and more accurate diagnoses and treatment decisions for genetic conditions.

End-to-End Bioinformatics Solution for Next-Generation Sequencing
How VarSome Clinical works?
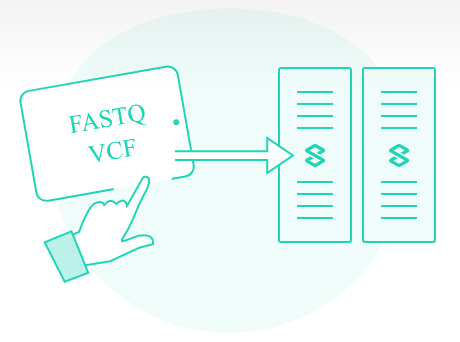
01 - How VarSome Clinical works?
VarSome Clinical accepts FASTQ and VCF files. You can upload the data easily and securely through its web interface or you can harness VarSome’s powerful API for an automated and secure data transfer. Once the data are uploaded, you can start the analysis!
With VarSome Clinical you can upload Illumina FASTQ files for variant discovery, annotation, and classification. You can perform analysis for multiple samples, such as risk carrier screening for couples, analysis of tumor-normal samples, studies of trios and families or comparative analysis of families of any size or small cohorts.
With VarSome Clinical you can upload a VCF file containing single or multiple samples (tracks) for variant annotation and classification. Subsequently, you can apply any number of sophisticated dynamic and/or algorithmic filters to quickly find the causative variants, or other variants of interest.
VarSome Clinical doesn’t call variants on all samples together to perform a joint analysis. We have developed a workflow that allows us to decouple the initial identification of potential variant sites from the genotyping step, which is the only part that really needs to be done jointly.
With VarSome Clinical, you can perform analysis for multiple samples, such as risk carrier screening for couples, analysis of tumor-normal samples, studies of trios and families or comparative analysis of families of any size or small cohorts.
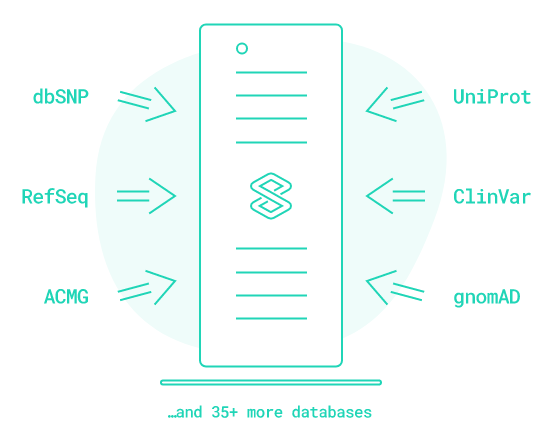
02 - Annotation & Classification
VarSome Clinical’s robust pipeline is capable of analyzing whole genomes, exomes, and gene panels, for individual samples, trios, families, and cohorts in minutes. VarSome Clinical leverages the massive cross-referenced knowledge base of the free VarSome and offers also access to licensed databases.
VarSome offers a massive cross-referenced knowledge base consisting of 35+ public genomic databases, representing over 33 billion data points, plus contributions from a 200’000-strong community. But there is more to it: whenever a public database is updated, VarSome processes it and makes it available for annotation and classification!
VarSome displays automated variant classification according to the guidelines of the American College of Medical Genetics and Genomics (ACMG). Each ACMG rule is explained, along with why it has been triggered, or why not. If you have some additional evidence, you can manually turn on or off other ACMG rules and easily reach the final verdict for your variant.
With VarSome Clinical you can access proprietary and licensed data sets of 3rd party data providers on a condition you possess corresponding license. You can also import your own local database with allele frequencies and integrate it privately in VarSome Clinical for annotation and classification of your variants.
VarSome Clinical comes with extensive Quality Control Report which covers pipeline settings as well as statistics for alignment, variant calling, ACMG and more. Gene Coverage Report and Coding Coverage Report is also available.

03 - Intuitive Web Portal
VarSome Clinical’s feature-rich and intuitive web interface allows filtering variants according to pathogenicity, ACMG classification, allele frequency, gene list or phenotype, to name a few! Dynamic and algorithmic filters allow you to perform simple or advanced filtering, like segregation analysis, identification of de novo variants or variants in imprinted genes, and much more!
Dynamic filters are combinations of conditions to apply to your samples in order to reduce your search space. They are powerful, yet easy to set up, modify and apply in seconds to any of your samples. You can filter variants based on allele frequency, pathogenicity, ACMG, zygosity, function or gene list, to name a few.
Algorithmic filters are sophisticated filters that can be fully customized according to your workflow and other specific needs. With algorithmic filters, you can perform segregation or gene list-based analysis, find compound heterozygous variants, identify de novo variants or variants in imprinted genes, to name a few.
VarSome Clinical allows you to build your own database of samples and variants. You can comment on variants and set up custom variant classifications to be shared with the members of your team. You may even share data at single variant level with your partner institutions on the condition that both parties consent in writing. With VarSome Clinical you can also import your own local database with allele frequencies and use it for variant annotation and classification.
On top of custom variant classifications and comments sharing with the members of your team you may even share data at single variant level with your partner institutions on the condition that both parties consent in writing.
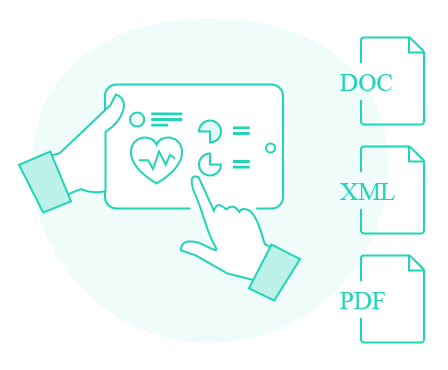
04 - Clinical Report Generation
Once you have narrowed down the list of variants of interest, you can proceed with the generation of the clinical report, which includes all the details of your variants, including literature references and your custom comments. The layout of the report can be fully customized according to your unique branding policy.
VarSome Clinical allows you to generate a clinical report containing the variants you choose for each sample. The report includes drug-related information (based on genes), literature references, and your custom comments.
VarSome Clinical is certified as an In-vitro Diagnostic Medical Device according to the requirements of EC 98/79/EEC. In addition, we are certified as a company with ISO 13485 for quality management system for medical devices.
VarSome Clinical comes with functionality to track the usage of the platform by users for auditing purposes.
Key Benefits
CE-IVD Mark
VarSome Clinical is certified as an In-vitro Diagnostic Medical Device according to the requirements of EC 98/79/EEC.
HIPAA Compliant
VarSome Clinical is compliant with HIPAA and HITECH Act, ensuring that we have the required safeguards in place to protect ePHI.
ISO 13485
We are certified as a company with ISO 13485 for quality management system for medical devices.
End-to-End Solution
VarSome Clinical provides validated and clinically certified genome-scale variant discovery, annotation and classification.
Variant Knowledge Base
Cross-referenced data from 35+ genomic databases plus contributions from a 200’000-strong community .
Programming Interface
VarSome Clinical comes with powerful API for automated data transfer in both directions.
Did you know?
Variant Discovery
VarSome Clinical’s variant discovery pipelines have been designed to achieve high quality standards, such as reproducibility, sensitivity and precision:
1. Sensitivity: 99.8% for SNVs and 99.5% for indels
2. Precision: 99.8% for SNVs and indels
Precision Contests
VarSome Clinical’s variant discovery rocks! It received top marks in the recent contest organized by the Food and Drug Administration, and it was steadily in the top ranks in all metrics, as in previous precision FDA contests. These results are a testament to the VarSome team’s total commitment to excellence.
Experts in Big Data
To date VarSome has aggregated and cross-referenced over 35 leading databases and other data resources, representing over 33 billion data points, and new ones are added constantly over time. But there is more to it – whenever a data resource is updated, VarSome quickly makes it available for annotation and classification of your variants!
Network of Geneticists
As VarSome Clinical has been deployed by dozens of institutional customers across many countries, you can benefit from VarSome’s extensive network of molecular geneticists and health care professionals who contribute to the identification of likely causal variants, along with a summary of available therapeutic options.
